The Human Microbiome
We are not alone. The human body harbors more bacterial cells than human cells, and approximately one kilogram of bacteria reside in the human gut. From the moment we are born, bacteria begin training our immune system to fight disease, and bacteria in our intestinal tract aid in digestion, releasing nutrients and vitamins for our use. Microbial imbalance has been linked do a wide range of disease states, including inflammatory bowel disease, colon and liver cancers, diabetes, autism, and obesity. However, the molecular mechanisms by which the microbiota affects human health are largely unknown.
The long-term goal of the Devlin lab is to understand and control the chemistry of human-associated bacteria in order to uncover how the microbiome affects human health and disease.
Our lab uses small molecules to study and manipulate human-associated bacteria in order to better understand how the microbiome affects human health and disease. The lab leverages expertise from different fields, including microbiology, biochemistry, analytical and organic chemistry, molecular and cellular biology, and germ-free mouse experiments. Project areas in the lab include:
Investigating bacterial metabolism of host-produced molecules and how these compounds affect host physiology
Host-produced compounds such as steroids and vitamins act as crucial signaling molecules and regulators of host biology, but their chemical transformations by gut bacteria have been relatively unexplored. We hypothesize that chemical modifications of host-produced molecules by gut bacteria produce metabolites with altered functions compared to the parent compounds. We aim to uncover both the ways in which bacteria make these molecules (that is, what bacterial genes and proteins are responsible) and the biological roles of these bacterially modified metabolites.
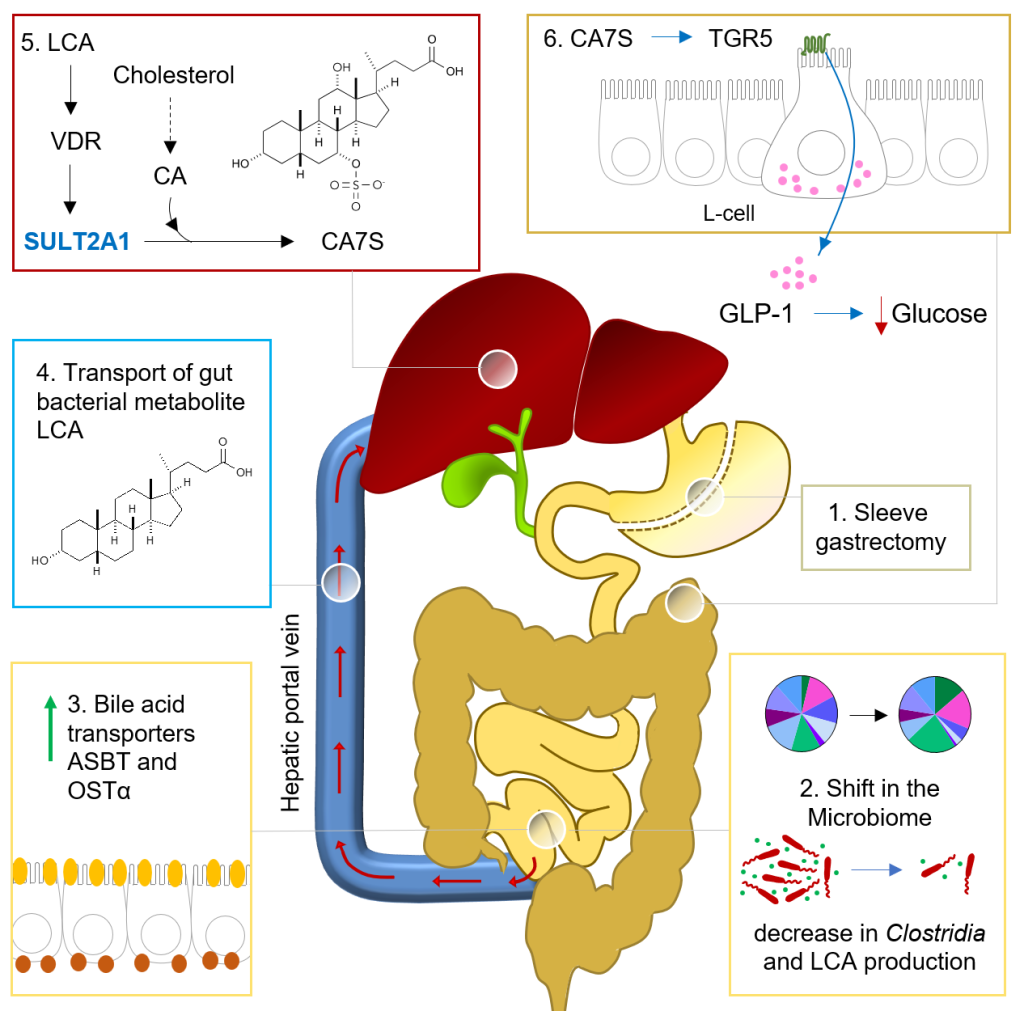
Bile acids are steroidal natural products that are synthesized from cholesterol in the liver and constitute an important part of the molecular environment of a healthy human gut. Bacteria in the large intestine transform human-derived primary bile acids into secondary bile acids in near-quantitative fashion. Both primary and secondary bile acids exert wide-ranging biological effects, from behaving as detergents that aid in digestion to acting as causative agents in colon and liver cancer to binding nuclear receptors and initiating downstream metabolic cascades. Despite their important role in human health, we know relatively little about which bacteria metabolize bile acids or which genes are responsible. By uncovering how and why bacteria transform these compounds, we will pave the way for the rational alteration of the human gut microbiome to treat diseases such as inflammatory bowel disease and obesity.
Representative publications:
- Bile acids affect T cell differentiation and function [Nature 2022] [Cell Host Microbe 2021a]
- Bile acids affect host metabolism [Nat. Chem. Biol. 2021] [Cell Host Microbe 2021b] [eLife 2018]
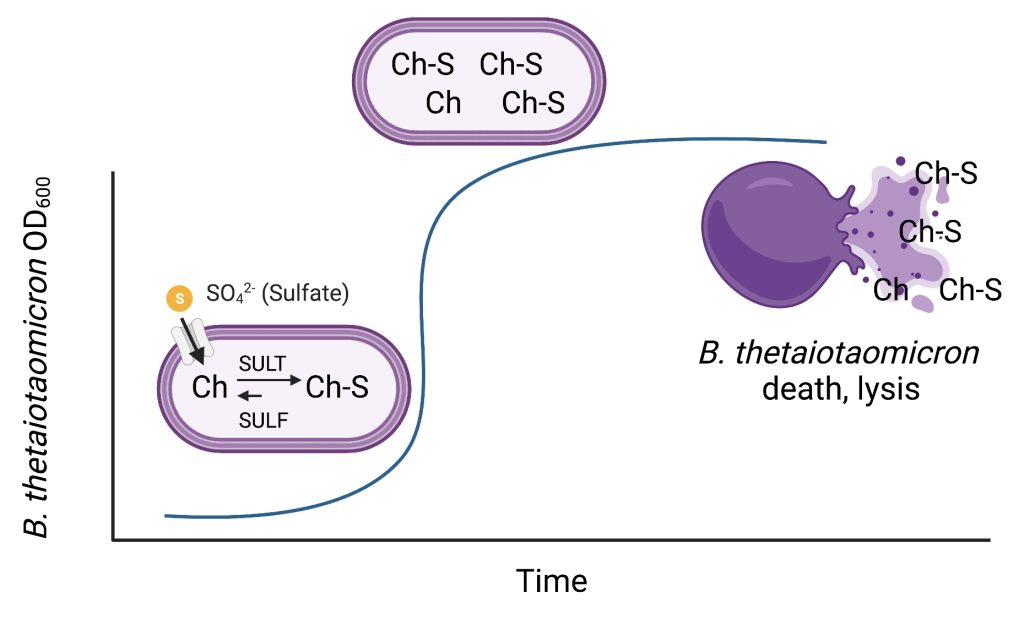
Steroids and sterols are potent signaling molecules that play essential roles in the regulation of fundamental biological processes, including metabolism and stress response, sexual differentiation and reproduction, and salt and water balance. Recent research indicates that bacterial interactions with steroids affect host stress response, immune function, and behavior. For example, introducing the human gut bacterium and probiotic Lactobacillus into mice reduces the levels of stress-induced corticosterone (the mouse equivalent of cortisol) and limits depression-like symptoms. These effects may be linked to immune cell function. Despite these intriguing correlative findings, the causal links between bacteria, steroidal hormones, and host systems remain elusive. We aim to uncover ways in which human gut bacteria affect host biology through the chemical modification of host-derived steroids.
Representative publication:
Controlling bacterial metabolism in vivo using small molecule inhibitors and probes of gut bacterial enzymes
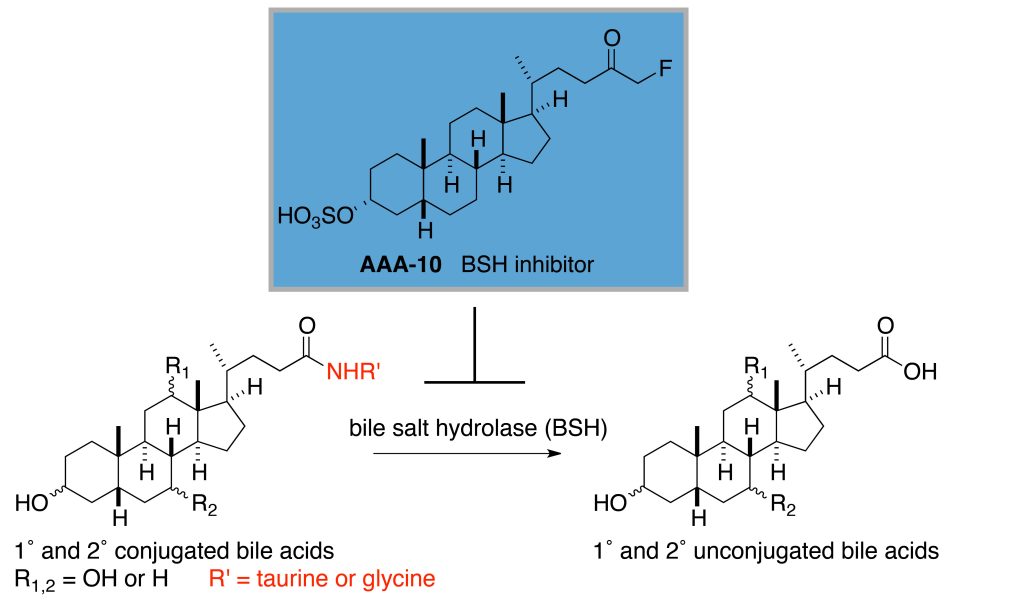
There is currently growing interest in targeting the gut microbiome for the betterment of human health and the treatment of disease. However, there are very few examples of interventions that selectively target the microbiome in vivo. Most treatments are like sledgehammers – they are either non-selective (e.g., antibiotics) or target a wide variety of bacterial metabolites (e.g., resins like cholestyramine). Moreover, from the point of view of discovery-based science, our ability to control the levels of bacterially produced small molecules in vivo in hosts containing a complete microbiome is currently limited. While germ-free animals are a useful tool, in order to investigate the effects of bacteria on the host in the context of the full complement of bacterial and mammalian processes, it is necessary to perform experiments in conventionally raised animals. In order to address the challenge of controlling bacterial metabolism in vivo and to pave the way for new therapeutic strategies to improve human health and treat disease, we are designing, synthesizing, and utilizing small molecule probes of gut bacterial enzymes.
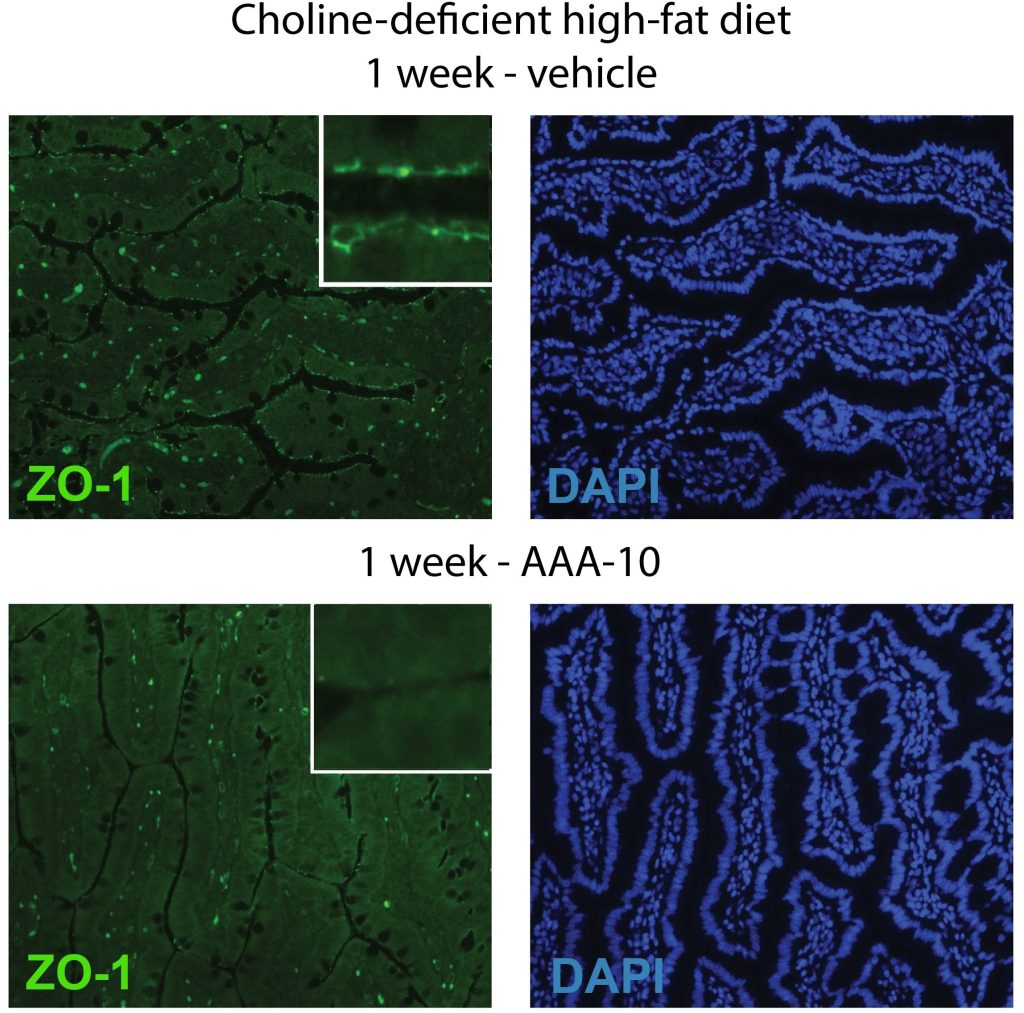
Recently, the lab has developed selective small molecules tools that act more like scalpels. That is, they specifically target one class of enzymes in gut bacteria, bile salt hydrolases (BSHs). Inhibition of these enzymes does not kill the bacteria but rather predictably alters the bacterial metabolism of bile acids. In doing so, a group of molecules that we have now shown are beneficial to the host by protecting gut barrier integrity are increased in vivo. Because intestinal barrier integrity is foundational for gut health, this work has implications for the prevention or treatment of a range of human diseases, including inflammatory bowel disease (IBD), liver disease, type 1 and type 2 diabetes, cardiovascular disease, and depression.
Representative publications:
